CytokineLink

CytokineLink collates cytokine interaction networks, describing how cytokines may affect one another.
What are cytokines?
Cytokines are small peptides that facilitate communication between various cell types and play a crucial role in the immune system.
Cytokine interactions
Cytokines can activate or inhibit the production and activity of other cytokines in target cells. These cytokine interactions help advance the immune response to infections and illnesses.
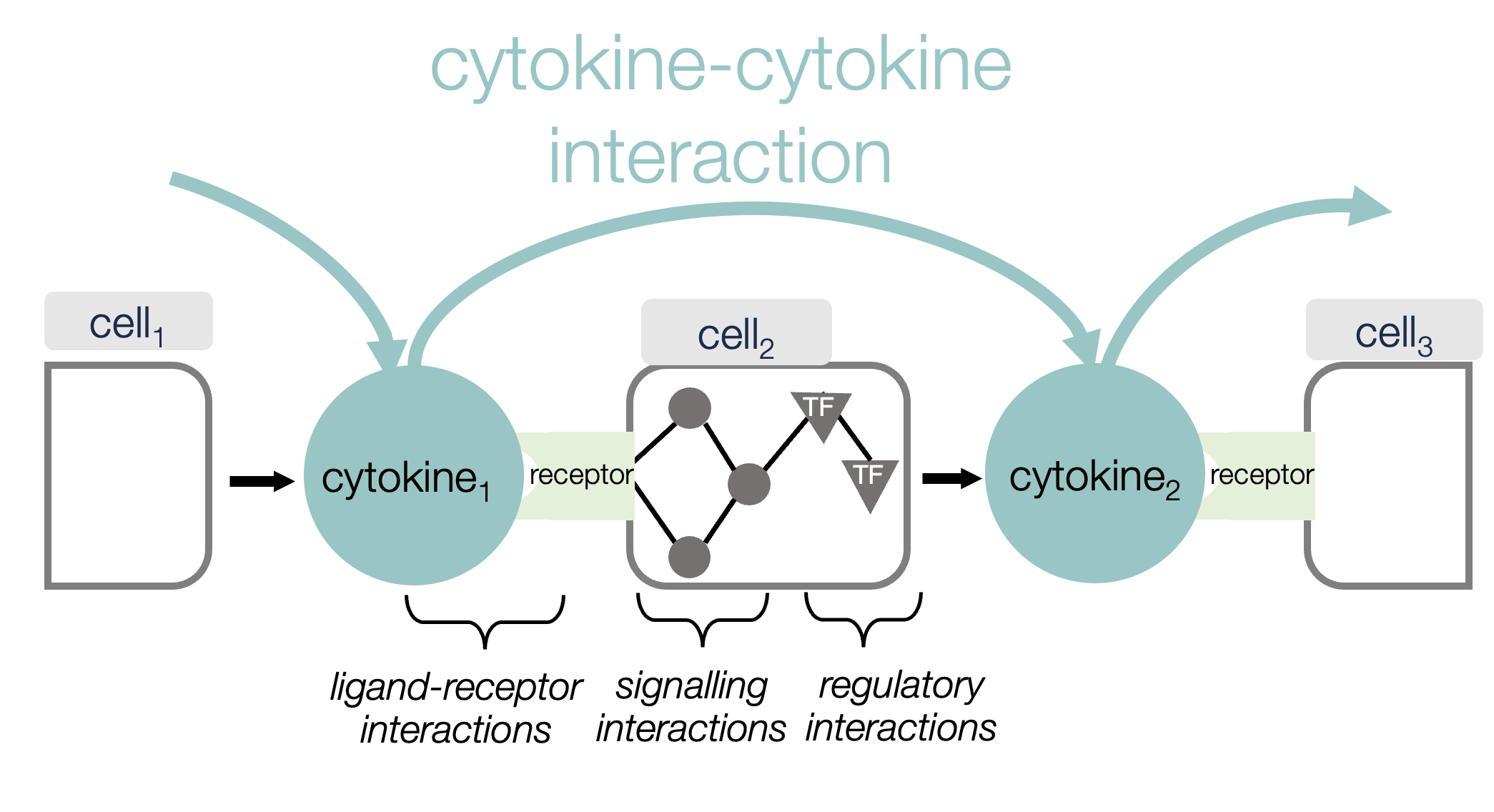
Why cytokine interactions?
By studying the broader network of cytokine interactions instead of the effects of individual cytokines we aim to map how the immune system shapes itself in disease. Modulating cytokine communication to manage an overactive immune response is often a key objective in drug development, and by understanding how cytokines influence each other we may offer alternative intervention points, and better understand why certain therapies do and don’t work.
Cytokine interaction resources
Cytokine network dysregulation in Ulcerative Colitis (2024)
In this study, we used single-cell transcriptomics data from colonic biopsies of immunotherapy treatment-naive and immunotherapy-treated ulcerative colitis (UC) patients to build cytokine networks characterising active cytokine pathways in UC. The networks effectively captured known physiologically relevant cytokine-cytokine interactions which could be recapitulated in vitro in UC patient-derived colonic epithelial organoids and highlighted novel facets of UC pathogenesis, including a cytokine subnetwork that is unique to treatment-naive UC patients.
Our preprint: Decoding Cytokine Networks in Ulcerative Colitis to Identify Pathogenic Mechanisms and Therapeutic Targets
The networks can be interactively accessed on the NDEx platform:
Cytokine interactions from the Immune Dictionary (2024)
This network contains the cytokine-cytokine interactions collated from the supplementary materials of Cui et al., 2023.
The network can be accessed via the NDEx network repository
Cytokine interactions from CytoSig (2023)
In this project, we created a network of cytokine interactions by processing transcriptomics data from over 2000 datasets deposited to the CytoSig database. By applying CytoSig’s significance analysis protocol, we identified statistically significant cytokine - cytokine interactions. The resulting network is a signed, directed map of cytokine communication, comprising 544 interactions among 155 cytokines. These interactions are annotated with the potential intracellular pathways connecting the cytokines and their respective stimulatory or inhibitory effects, based on the sum of deposited expression data.
The network can be accessed via the NDEx network repository
CytokineLink (2021)
In our earlier work, we collated the most prevalent cytokines from the literature and assigned the proteins and their corresponding receptors to source tissue and blood cell types based on enriched consensus RNA-Seq data from the Human Protein Atlas database. To assign more confidence to the interactions, we integrated the literature information on cell–cytokine interactions from two systems of immunology databases, immuneXpresso and ImmunoGlobe.
The network can be accessed via the NDEx network repository